Track 1: White Matter Multiple Sclerosis Lesions Segmentation
Task
The task of this track is the segmentation of White Matter Lesions (WML) of Multiple Sclerosis (MS) in Magnetic Resonance Images (MRIs). This involves the generation of a 3D per-voxel segmentation mask identifying each voxel as lesion or non-lesion tissue [Rovira et al., 2015][Wattjes wt al., 2021].
MRIs are multi-modal images of the brain, with MS diagnosis being based mainly on :
- T1-weighted
- FLAIR (Fluid-Attenuated Inversion Recovery)
The objectives of this track are double-sided, as the submission will be evaluated on their:
- Voxel-scale lesion segmentation performance
- Quality of voxel-scale uncertainty estimates to handle the domain shifts in the dataset
Data
The dataset includes the following datasets:
- MSSEG-1 [Commowick et al., 2018],
- ISBI [Carass et al., 2017],
- PubMRI [Lesjak et al., 2017]
- Lausanne (private, not released for privacy reasons), provided by the Swiss universities of Lausanne and Basel
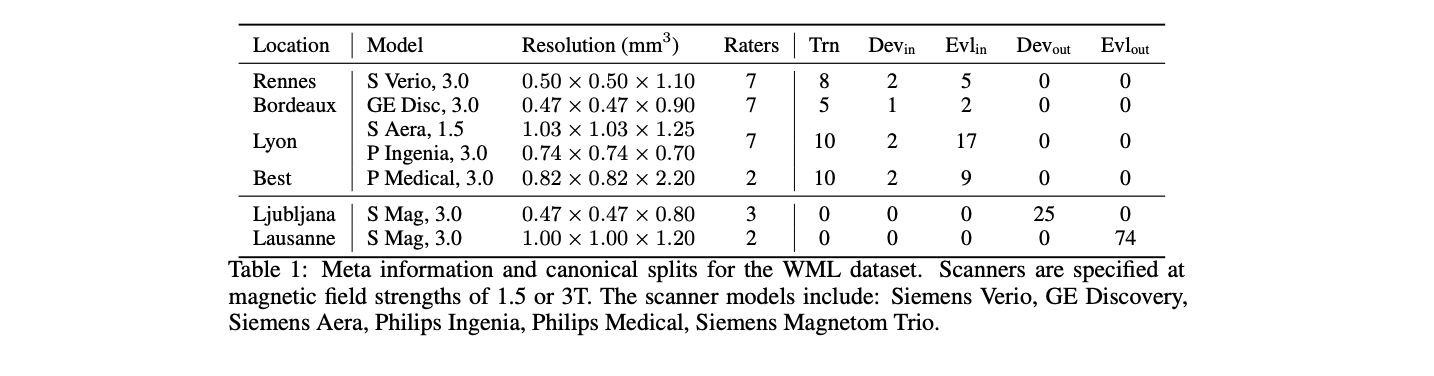
The private dataset is used for the external evaluation of the challenge submissions. A detailed summary of the provenance of the patient scans, scanner types, magnetic field strengths and original image resolution is given in Table 1.
Data preprocessing and splits
The supplied data have already undergone our preprocessing and do not need further preprocessing steps.
Our preprocessing includes de-noising, skull-stripping after registering the T1-weighted to FLAIR, bias field correction and interpolation to a 1mm iso-voxel space. The ground truth masks are also interpolated to the 1mm iso-voxel space and are obtained as a consensus of multiple expert annotators (and as a single mask for Best and Lausanne, which have 2 annotators only) .
The data is split into:
- in-domain splits Trn (training), Evl_in (evaluation) and Dev_in (development)
- out-of-domain shifted splits Dev_out (shifted development) and Evl_out (shifted evaluation)
```
Zenodo ├── Best/ │ ├── Trn/ │ │ ├── FLAIR/ │ │ ├── gt/ │ │ ├── fg_mask/ │ │ └── T1/
│ ├── Dev_in/ │ └── Evl_in/ ├── Ljubljana/ │ └── Dev_out/ └── MSSeg/ ├── Trn/ ├── Dev_in/ ├── Evl_in/ └── unlabelled/ └── FLAIR/
```
Data organisation. First level directories represent the devision by the source datasets. Second level directories represent the splits. To obtain the full training set combine data from ISBI/Trn/ and MSSeg/Trn. The third level directories contain the modalities: FLAIR, gt (ground truth), fg_mask (foreground mask), T1 (T1-weighted).

Example of the input data and expected outputs. The first row shows a training 3D FLAIR scan and its ground truth binary mask of the lesions (in green). The second row shows the predicted mask by our baseline model (in red). The last row illustrates the uncertainty heatmap for the baseline predictions computed with reverse mutual information. High uncertainty regions are located on the borders of lesions.
Evaluation
MS lesion segmentation of 3D MRI images is typically assessed via the Dice Similarity Coefficient (DSC) [Dice, 1945; Sorensen et al., 1948] between manual lesion annotations and the model's prediction. However, DSC is strongly correlated with lesion load - patients with higher lesion load (volume occupied by lesion) will have a higher DSC [Reinke et al., 2021].
We will evaluate:
- The lesion segmentation by the normalized Dice Similarity Coefficient (nDSC). This version, compared to the original [Dice, 1945; Sorensen et al., 1948], corrects for the systematic bias between manual lesion annotations and the model's prediction. More info about nDSC.
- Error-retention curves on the foreground voxels to assess the quality of the uncertainty estimation as the area between the curve and a horizontal line at 1 that corresponds to high nDSC. More info about the error-retention curves.
Baseline
We provide a 3D UNET baseline based on the work by [La Rosa et al., 2020].
The model is trained for 300 epochs with early-stopping on Dev-in. At training time, 32 patches of 96x96x96 voxels are sampled from the centre of a lesion in the input volume. At inference time, overlapping patches (by 25%) are selected across the whole 3D volume. Gaussian weighted averaging is used for the final prediction of each voxel belonging to multiple patches.
The segmentation map is obtained by thresholding the predicted probabilities by a threshold tuned on Dev-in. Deep ensembles are formed by averaging the output probabilities of 3 distinct models trained with different seed initialisation.
As each single model yields a per-voxel probabilistic prediction, ensemble-based uncertainty measures [Malinin, 2019; Malinin & Gales] are available for uncertainty quantification. Our ensembled models use reverse mutual information [Malinin et al., 2021] as the choice of uncertainty measure.
Get started
If you want to get a quick idea about how to handle new medical imaging data format, you will find useful a GitHub dedicated to the task: GitHub. There you will be able to find code for handling the data format, code to reproduce the baseline model and code of evaluation metrics.
Ready to start? Download the data from Zenodo and start creating your cool model for white matter lesion segmentation.
Ready to submit? Visit the submission page explaining how to build and submit you docker model.
